If you are performing computationally intense calculations, we recommend using ChemApp compared to Equilib, especially when iterating over a lot of conditions. Due to the single threaded nature of ChemApp, each run of ChemApp only employs a single CPU core. Most, if not all current computer systems have multiple CPU that should be employed if running computationally intense calculations. Python itself is relatively slow in execution, compared to many other, especially non-interpreted programming languages. Therefore, if working with larger datasets, better utilization of the available resources is almost mandatory.
Before attempting parallelization, a key question surrounding multiprocessing comes from identifying bottlenecks and the ‘weakest link’ in the full sequence of calculations. With respect to ChemApp, and the Python modules that GTT supplies to interface it with Python, this question cannot be answered generally and depends on the problem at hand, meaning both the thermodynamic system that the calculations are performed on, as well as the conditions that are used.
A few best practices to check when trying to improve the calculation speed are:
- Try to use the minimum number of selected phases. Including unstable phases that are never stabilizing throughout the calculation set are wasting both memory and computational resources for stability checks.
- The selection of multiple phase copies (Option I, J in Equilib, and having
Status.ENTEREDfor<phase>#1,<phase>#2(and<phase>#3)) can yield quite extensive computational penalties for numerical reasons. If possible, you could try to reduce the number of copies by setting the status of<phase>#2(or #3) toStatus.ELIMINATED. Be cautious though, as for many phases and conditions, multiple copies are mandatory to achieve correct results. It requires understanding the thermodynamic system to make the right choice. - Some phase models, especially the more intricate and complex solution phase models that perform computationally demanding phase-internal equilibration, such as the quasichemical model or magnetic sublattice site-fraction ordering are significantly more expensive to calculate for ChemApp. Often, it may not be possible to avoid these phases. However, sometimes these can be adequately replaced by some less detailed model.
- Remove unused system components. If your calculation contains a system component for which the amount is zero, consider removing it. Even if set to zero, the phase constituents and compounds that contain that system component are added to the calculation base and may considerably increase the size and complexity of the loaded thermodynamic system.
- Generally, target calculations are iteratively calculating to achieve the target calculation. Depending on your problem space, reformulating the intention of your calculations into single point calculations may yield a lower calculation time, especially if the target calculations are prone to fail, e.g. because the conditions set make the target unreachable.
- When using Equi2Py, the generated code typically fetches the
EquilibriumCalculationResult(orStreamCalculationResult) object from a calculation. Generating these objects has some computational cost compared to only retrieving specific values from a calculation result. In addition, much less memory is needed, if a result object does not need to be stored.
However, all of the above will and should not deter you from trying to maximize the efficiency of your provided computational resources. Since this article is not targeting your general understanding of computational efficiency, different type of parallelization or processing will not be deeply explained, but a short rundown of an almost always applicable technique will be given.
Python is peculiar as a language for parallel processing due to the Global Interpreter Lock. One can think of the Global Interpreter Lock (GIL) in Python like a “talking stick” in a group conversation. Only the person holding the talking stick can speak, and others have to wait their turn. Similarly, the GIL ensures that only one part of your Python program can run at a time, preventing conflicts. While this helps avoid chaos, it can also slow things down if you have many parts that want to “speak” or run simultaneously. This restriction very deeply affects the ways to achieve parallelism.
In Python, multiprocessing and threading, with equivalently named packages multiprocessing and threading from the standard library can be used. A brief summary of their characteristics and differences can be found in Table 1:
| Python Package Name | Multiprocessing | Threading |
|---|---|---|
| Concept | Processes run in separate memory spaces and have their own interpreter and Python Global Interpreter Lock (GIL). This makes multiprocessing suitable for CPU-bound tasks that can benefit from parallelism. | Threads run in the same memory space, sharing data, and are suitable for I/O-bound tasks where a program is waiting for external events (e.g., network communication, file I/O). |
| Global interpreter lock | Each process has its own GIL, enabling true parallelism for CPU-bound tasks. Multiple processes can run simultaneously on multiple CPU cores. | One thread to execute Python bytecode at a time in a single process. This makes it challenging to achieve true parallelism in CPU-bound tasks. |
| Memory | Processes have separate memory spaces, and communication between them usually involves inter-process communication (IPC) mechanisms, such as queues or pipes. | Threads share the same memory space, which can make it easier to share data between threads but also requires synchronization mechanisms (e.g., locks) to prevent data corruption. |
| Runtime Complexity | Involves more overhead due to separate processes, but it’s more suitable for CPU-bound tasks and provides better parallelism, if the processes are not memory or I/O bound. | Has lower overhead compared to multiprocessing, but you need to be cautious about thread safety and race conditions when sharing data between threads. |
| Summary | Use for CPU-bound tasks that can benefit from parallelism with smaller memory needs. | Use for I/O bound tasks and situations where simplicity and data sharing are essential |
For your application that uses ChemApp, both options may be chosen and can even be combined. However, ChemApp itself is a single threaded process, and the singleton programming pattern being used to implement the connection between the ChemApp library and the Python interface blocks threading for your ChemApp calculations. It is still possible – and potentially reasonable, to separate your program into ChemApp- and non-ChemApp-bound code and threads. However, this depends mostly on application code unrelated to ChemApp and is not suitable for this article.
Therefore, the following part will focus on quick and easy multiprocessing approaches for cases where multiple independent ChemApp calculations are run in parallel.
Generally, code optimization strategies should not be implemented blindly – always profile your code first, to figure out bottlenecks and inefficiencies. Do not assume that the following example is a cookiecutter for every processing script.
Example
We will start with a simple ChemApp script (of a “costly” calculation), using 11 elements in a combination of 12 phase constituents of a steel/slag system, at varying temperatures. The calculation is relatively meaningless, so don’t bother to make thermochemical sense of the script. Despite that, if you are willing to follow along, the corresponding Equilib file is TODO.
As a baseline, the Equilib calculation which calculates 6 equilibria at varying temperatures takes roughly 4 minutes on my test machine, which means about 40 seconds per individual point. Running the Equi2Py converted script on my computer takes a bit less time. That in itself is not the performance improvement we are looking for.
Note: Usually, I would recommend working with Jupyter notebooks to be able to quickly observe results. However, using multiprocessing in Jupyter notebooks is a bit more intricate than in pure Python. Therefore, we will only use ‘bare’ Python scripts.
Find the code here on GitHub.
#> Initialization and general setup
import os
from pathlib import Path
import numpy as np
from collections import OrderedDict
import chemapp as ca
from chemapp.friendly import (
Info,
StreamCalculation as casc,
ThermochemicalSystem as cats,
EquilibriumCalculation as caec,
Units as cau,
TemperatureUnit,
PressureUnit,
VolumeUnit,
AmountUnit,
EnergyUnit,
Status,
)
from chemapp.core import (
LimitVariable,
ChemAppError,
)
#> Load data file
cst_file = Path("SlagHeating.cst")
# load the database file
cats.load(cst_file)
#> Units setup
# It is possible to change the units before retrieving results, or in between making multiple inputs. However, mixed
# unit input is currently not generated automatically.
cau.set(
T = TemperatureUnit.C,
P = PressureUnit.atm,
V = VolumeUnit.dm3,
A = AmountUnit.gram,
E = EnergyUnit.J
)
#> Fixed conditions
# setting general boundary conditions of the calculation(s)
casc.set_eq_P(1.0)
#> Stream setup
# Create streams. The names are defaults of "#1", "#2", ...
# Feel free to make these more reasonable, but take care to edit everywhere they are used
casc.create_st(name="#1", T=25, P=1)
#> Variable conditions
# original condition for T was 1500.0, which would not include 1500.0.
# Increasing it by 20.0 yields the inclusive upper boundary.
T_range = np.arange(1400.0, 1520.0, 20.0)
# we keep the results in an OrderedDict, to access by variable value
# if wanted, the .values() attribute of results can be easily used like a list.
results = OrderedDict()
#> Calculation
for T in T_range:
# set incoming amounts
# since this is a calculation with initial conditions, input needs to be made
# using phase constituents
casc.set_IA_pc("#1", "Al2O3_gamma(s)", "Al2O3_gamma(s)", 15.0)
casc.set_IA_pc("#1", "CaO_Lime(s)", "CaO_Lime(s)", 33.0)
casc.set_IA_pc("#1", "MgO_periclase(s)", "MgO_periclase(s)", 15.0)
casc.set_IA_pc("#1", "SiO2_Quartz(l)(s)", "SiO2_Quartz(l)(s)", 37.0)
casc.set_IA_pc("#1", "FeO_Wustite(s)", "FeO_Wustite(s)", 0.001)
casc.set_IA_pc("#1", "Fe2O3_hematite(s)", "Fe2O3_hematite(s)", 0.001)
casc.set_IA_pc("#1", "Cr2O3_solid(s)", "Cr2O3_solid(s)", 0.001)
casc.set_IA_pc("#1", "MnO_solid(s)", "MnO_solid(s)", 0.001)
casc.set_IA_pc("#1", "TiO2_Rutile(s)", "TiO2_Rutile(s)", 0.001)
casc.set_IA_pc("#1", "K2O_solid(s)", "K2O_solid(s)", 0.001)
casc.set_IA_pc("#1", "Na2O_Solid-A(s)", "Na2O_Solid-A(s)", 0.001)
casc.set_IA_pc("#1", "V2O5_solid(s)", "V2O5_solid(s)", 0.001)
try:
# calculate
casc.calculate_eq()
#fetch results
results[T] = casc.get_result_object()
except ChemAppError as c_err:
print(f"[<T> = { T }] ChemApp Error {c_err.errno}:")
print(c_err)
# change this if you want to capture a calculation error
# maybe input a dummy value (or other, adequate replacements?):
# results[T] = None
pass
print("End of the Python script.")
Code language: Python (python)There are multiple ways how to instrument the multiprocessing package (Python Library Documentation). As a demonstration, we will use the relatively high abstraction that multiprocessing.Pool provides – which removes the necessity to explicitly start, stop or interact with processes and which manages the interprocess communication. This is possible since the type of multiprocessing that we are implementing does not use any ‘hidden’ shared resources that we need to synchronize during the multiprocessing part of the execution. multiprocessing.Pool basically spawns a number of processes that are running completely independently and only return the results at the end of their respective run.
To employ Pool, the calculation part of the script needs to be adapted to use a calculation function, as that is called from the workers and parametrized for the values varied between runs. Additionally, since the worker will load and process the code file in a mostly independent way, the name-main idiom has to be implemented, so that the Pool mechanism can keep track of the main process and respective child processes.
The changes made:
- Importing the necessary
Poolclass frommultiprocessing. - Wrapping the calculation code into a function called
calculate, which is parametrized for the variable temperature. - The
calculateroutine returns the result object, so that it can be collected in the main process. - Guard the ‘iterative’ calculation by using the
__name__ = "__main__"idiom. - Using the
Poolcontext manager access to the pool of available processes. - Using the
mapfunctionality to distribute the calculation of the routine. - Some simple timing code using
time.perf_counter_ns()has been added.
Find the code here on GitHub.
from pathlib import Path
import numpy as np
from collections import OrderedDict
from multiprocessing import Pool
import time
from chemapp.friendly import (
StreamCalculation as casc,
ThermochemicalSystem as cats,
Units as cau,
TemperatureUnit,
PressureUnit,
VolumeUnit,
AmountUnit,
EnergyUnit,
)
from chemapp.core import (
ChemAppError,
)
# > Load data file
cst_file = Path("SlagHeating.cst")
# load the database file
cats.load(cst_file)
# Units setup
cau.set(
T=TemperatureUnit.C,
P=PressureUnit.atm,
V=VolumeUnit.dm3,
A=AmountUnit.gram,
E=EnergyUnit.J,
)
# Calculation
def calculate(T: float):
"""Calculate equilibrium at a given temperature"""
# print(f"Process {os.getpid():>6d}: calculating at T =", T)
# Fixed conditions
casc.set_eq_P(1.0)
# Stream setup
casc.create_st(name="#1", T=25, P=1)
# Set incoming amounts
casc.set_IA_pc("#1", "Al2O3_gamma(s)", "Al2O3_gamma(s)", 15.0)
casc.set_IA_pc("#1", "CaO_Lime(s)", "CaO_Lime(s)", 33.0)
casc.set_IA_pc("#1", "MgO_periclase(s)", "MgO_periclase(s)", 15.0)
casc.set_IA_pc("#1", "SiO2_Quartz(l)(s)", "SiO2_Quartz(l)(s)", 37.0)
casc.set_IA_pc("#1", "FeO_Wustite(s)", "FeO_Wustite(s)", 0.001)
casc.set_IA_pc("#1", "Fe2O3_hematite(s)", "Fe2O3_hematite(s)", 0.001)
casc.set_IA_pc("#1", "Cr2O3_solid(s)", "Cr2O3_solid(s)", 0.001)
casc.set_IA_pc("#1", "MnO_solid(s)", "MnO_solid(s)", 0.001)
casc.set_IA_pc("#1", "TiO2_Rutile(s)", "TiO2_Rutile(s)", 0.001)
casc.set_IA_pc("#1", "K2O_solid(s)", "K2O_solid(s)", 0.001)
casc.set_IA_pc("#1", "Na2O_Solid-A(s)", "Na2O_Solid-A(s)", 0.001)
casc.set_IA_pc("#1", "V2O5_solid(s)", "V2O5_solid(s)", 0.001)
try:
# calculate
casc.calculate_eq()
# return result
return casc.get_result_object()
except ChemAppError as c_err:
print(f"[<T> = { T }] ChemApp Error {c_err.errno}:")
print(c_err)
# This is a 'fallthrough' return, which is only reached if the calculation
# fails. Likely, this is not what you want.
return None
# this is the name-main idiom, which makes sure that the code is only executed
# when the script is run directly, not when it is imported, and which allows
# multiprocessing to work
if __name__ == "__main__":
results = OrderedDict()
# we just aim for some amount of calculations to be done
# using linspace allows to set the number of calculations
number_of_calculations = 80
T_range = np.linspace(1400.0, 1500.0, number_of_calculations)
# Set the number of processes to use
number_of_processes = 8
# Set the chunksize (number of calculations per process)
chunksize = 1
# start the timer
tstart = time.perf_counter()
print(f"Using {number_of_processes} processes")
# run the calculation, distributed on the number of processes
with Pool(processes=number_of_processes) as p:
results = p.map(calculate, T_range, chunksize=chunksize)
# print the timing result
print(f"{time.perf_counter() - tstart:.2f}s runtime")
# continue with your work with the results
# ...
print("End of script.")
Code language: Python (python)What the multiprocessing module does, is, roughly, to run the function (and imports!) on each process. It forks the child processes from the main process which gives somewhat a ‘headstart’ in that it does not have to initiate everything separately, but e.g. the copied instantiation of the child processes means that each ChemApp process that is being used in each child process is run independently of the other processes.
Of note, the supervising main process is collecting all the results into one OrderedDict, as before. This works because of the context wrapper ‘magic’ that is being used, which prevents race conditions and implements the proper inter-process communication. This could be done manually and may be mandatory in some parallelisation implementations, but again, this is outside the scope of this simple example.
multiprocessing.Pool certainly isn’t the only possible solution to using multiple processes. There are other components of the multiprocessing module, such as a Queue (for managing task distribution between processes, see Python Standard Library Documentation) and even async performers, which may be suitable if the workload cannot be as easily ‘anticipated’ as it is currently with the fixed number of temperatures over which to iterate. So if adaptive coarsening or dynamic calculation schemes are used, a Queue may be more robust and better at distributing the load.
There are also multiple different ways to distribute the load for a Pool. map distributes systematically and evenly, but other strategies are available. Please refer to the documentation for more information.
Results
On my 16 core machine, the parallelization yields perfectly expected results. I ran benchmarks on the calculation given above, averaging each run over 5 times to even out the inherent randomness coming with multitasking operating systems. The number of total calculations was fixed at 80 points – no particular reason for this, as this is purely for benchmarking the multiprocessing.
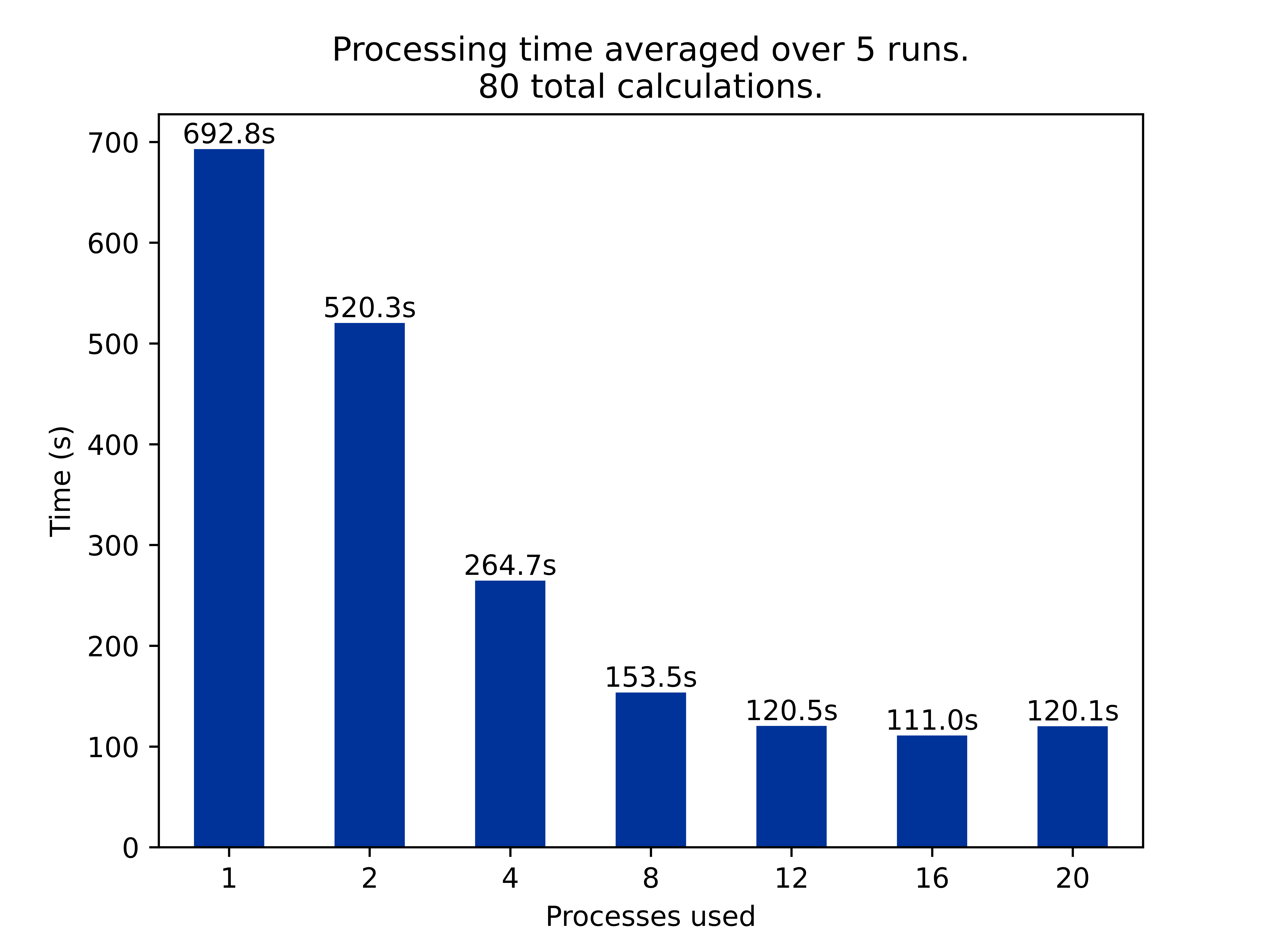
As a baseline, the calculation takes 692s, almost 11.5 minutes on a single core, whereas parallel execution on 16 cores takes less than 2 minutes, 1:50. At large, improvements scale as expected with increased number of processes, until the saturation of available ressources sets in – especially the switch from 12 to 16 processes does not yield much improvement, indicating that my work on this article and multitasking with various other open programs and computer loads take some ressources, too. As to be expected, more than 16 parallel processes can not increase performance, as the machine can’t run more concurrent processes, and the overhead of process creation and context switching for the operating system incur some cost. Of note, power managing/throttling logic of the Ryzen 7 Processor used here mean that single- and ‘few-‘ core utilization of the CPU will run at a higher clock than a fully saturated CPU.
Note: There possibly are benefits in having a larger process pool, as theoretically memory alignment and I/O optimizations of the operating system may improve the calculations. This is far beyond the reach of this article.
One additional consideration worth exploring is a more active adjustment of the amount of work that each process gets distributed – there is a parameter called chunksize for the map() routine. The relatively bare ChemApp calculation probably has very low loading overhead, given that the setup routines for a ChemApp calculation are computationally cheap, and that the loading of the Python runtime will be optimized. However, for two runs with 8 processes and a chunksize of 1 and 8, the results are 121.92s and 169.71s respectively. As surprising as that may sound, larger chunks do have a detrimental effect, possibly because a single child process may get ‘stuck’ for longer on a specific set of slow ChemApp calculations, as was observable from the CPU load. Again, the effect of this adjustment very strongly depends on your other computational costs around the ChemApp calculation, and whether you expect a large randomness of individual ChemApp calculation runtimes, which may occur e.g. in cases of target calculations. Other hardware properties of your computer may affect this also greatly.
Final words
Parallelization can be very powerful, and the results confirm this for ChemApp scripts written in Python. A near perfect improvement of the total calculation run time is possible with making use of the available parallel ressources. However, the possibilities and achievable speed-ups depend a lot on those components not explored in this post – may it be data intake, preprocessing or in the case of multi-step calculation, synchronization.
Feel free to contact us with your specific case. We are very willing to help!
The code used in this example and may more can be downloaded from the public examples GitHub repository:
https://github.com/GTT-Technologies/ChemApp-Examples
